Strategies¶
Summit has several machine learning strategies available for optimisation, as well as some more naive ones.
All strategies have a similar API. They are instantiated by passing in a Domain. New reaction conditions are requested using the suggest_experiments method which, potentially, takes results from previous reactions.
Contents
Bayesian Optimisation¶
Bayesian optimisation (BO) is an efficient way to optimise a wide variety of functions, inculding chemical reactions. In BO, you begin by specifying some prior beliefs about your functions. In many cases, we start with an assumption that we know very little. Then, we create a probabilistic model that incorporates this prior belief and some data (i.e, reactions at different conditions), called a posterior. In reaction optimisation, this model will predict the value of an objective (e.g., yield) at particular reaction conditions. One key factor is that these models are probabalistic, so they do not give precise predictions but instead a distribution that is sampled.
With the updated model, we use one of two classes of techniques to select our next experiments. Some BO strategies optimise an acquisition function, which is a function that takes in the model parameters and some suggested next experiement and predicts the quality of that experiment. Alternatively, a deterministic function can be sampled from the model, which is then optimised.
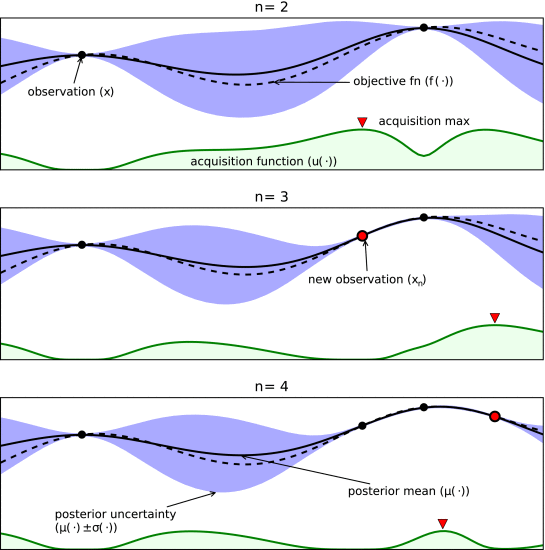
Illustration of how acquisition functions eanble BO strategies to reduce uncertainty and maximise objective simulataneously. Dotted line is actual objective and solid line is posterior of surrogate model. Acquisition function is high where objective to be optimal (exploration) and where there is high uncertainty (exploitation). Adapted from Shahriari et al.¶
To learn more about BO, we suggest reading the review by Shahriari et al.
The BO strategies available in Summit are:
TSEMO¶
-
class
summit.strategies.tsemo.TSEMO(domain, transform=None, **kwargs)[source]¶ Thompson-Sampling for Efficient Multiobjective Optimization (TSEMO)
TSEMO is a multiobjective Bayesian optimisation strategy. It is designed to find optimal values in as few iterations as possible. This comes at the price of higher computational time.
- Parameters
domain (
Domain) – The domain of the optimizationtransform (
Transform, optional) – A transform object. By default no transformation will be done on the input variables or objectives.use_descriptors (bool, optional) – Whether to use descriptors of categorical variables. Defaults to False.
kernel (
Kern, optional) – A GPy kernel class (not instantiated). Must be Exponential, Matern32, Matern52 or RBF. Default Exponential.n_spectral_points (int, optional) – Number of spectral points used in spectral sampling. Default is 1500. Note that the Matlab TSEMO version uses 4000 which will improve accuracy but significantly slow down optimisation speed.
n_retries (int, optional) – Number of retries to use for spectral sampling iF the singular value decomposition fails. Retrying chooses a new Monte Carlo sampling which usually fixes the problem. Defualt is 10.
generations (int, optional) – Number of generations used in the internal optimisation with NSGAII. Default is 100.
pop_size (int, optional) – Population size used in the internal optimisation with NSGAII. Default is 100.
Examples
>>> from summit.domain import * >>> from summit.strategies import TSEMO >>> from summit.utils.dataset import DataSet >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> columns = [v.name for v in domain.variables] >>> values = {("temperature", "DATA"): 60,("flowrate_a", "DATA"): 0.5,("flowrate_b", "DATA"): 0.5,("yield_", "DATA"): 50,("de", "DATA"): 90} >>> previous_results = DataSet([values], columns=columns) >>> strategy = TSEMO(domain) >>> result = strategy.suggest_experiments(5, prev_res=previous_results)
Notes
TSEMO trains a gaussian process (GP) to model each objective. Internally, we use GPy for GPs, and we accept any kernel in the Matérn family, including the exponential and squared exponential kernel. See [Rasmussen] for more information about GPs.
A deterministic function is sampled from each of the trained GPs. We use spectral sampling available in pyrff. These sampled functions are optimised using NSGAII (via pymoo) to find a selection of potential conditions. Each of these conditions are evaluated using the hypervolume improvement (HVI) criterion, and the one(s) that offer the best HVI are suggested as the next experiments. More details about TSEMO can be found in the original paper [Bradford].
The number of spectral points is the parameter that most affects TSEMO performance. By default, it’s set at 1500, but increase it to around 4000 to get the best performance at the cost of longer computational times. You can change it using the n_spectral_points keyword argument.
The other two parameters are the number of generations and population size used in NSGA-II. Increasing their values can improve performance in some cases.
References
- Rasmussen
C. E. Rasmussen et al. Gaussian Processes for Machine Learning, MIT Press, 2006.
- Bradford
E. Bradford et al. “Efficient multiobjective optimization employing Gaussian processes, spectral sampling and a genetic algorithm.” J. Glob. Optim., 2018, 71, 407–438.
-
suggest_experiments(num_experiments, prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using TSEMO
- Parameters
num_experiments (int) – The number of experiments (i.e., samples) to generate
prev_res (
DataSet, optional) – Dataset with data from previous experiments. If no data is passed, then latin hypercube sampling will be used to suggest an initial design.
- Returns
next_experiments – A Dataset object with the suggested experiments The lengthscales column tells the significance of each variable (assuming automatic relevance detection is turned on, which it is in Botorch). Smaller values mean significant changes in output happen on a smaller change in the input, suggesting a more important input. The variance column scales the output of the posterior of the kernel to the correct scale for your objective The noise column is the constant noise in outputs (e.g., assumed uniform experiment error)
- Return type
DataSet
SOBO¶
-
class
summit.strategies.sobo.SOBO(domain, transform=None, gp_model_type=None, acquisition_type=None, optimizer_type=None, evaluator_type=None, **kwargs)[source]¶ Single-objective Bayesian Optimization (SOBO)
This is a general BO method since it is a wrapper around GPyOpt.
- Parameters
domain (
Domain) – The Summit domain describing the optimization problem.transform (
Transform, optional) – A transform object. By default no transformation will be done on the input variables or objectives.gp_model_type (string, optional) – The GPy Gaussian Process model type. See notes for options. By default, gaussian processes with the Matern 5.2 kernel will be used.
use_descriptors (bool, optional) – Whether to use descriptors of categorical variables. Defaults to False.
acquisition_type (string, optional) – The acquisition function type from GPyOpt. See notes for options. By default, Excpected Improvement (EI).
optimizer_type (string, optional) – The internal optimizer used in GPyOpt for maximization of the acquisition function. By default, lfbgs will be used.
evaluator_type (string, optional) – The evaluator type used for batch mode (how multiple points are chosen in one iteration). By default, thompson sampling will be used.
kernel (
kern, optional) – The kernel used in the GP. By default a Matern 5.2 kernel (GPy object) will be used.exact_feval (boolean, optional) – Whether the function evaluations are exact (True) or noisy (False). By default: False.
ARD (boolean, optional) – Whether automatic relevance determination should be applied (True). By default: True.
standardize_outputs (boolean, optional) – Whether the outputs should be standardized (True). By default: True.
Examples
>>> from summit.domain import * >>> from summit.strategies import SOBO >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += CategoricalVariable(name='flowrate_a', description='flow of reactant a in mL/min', levels=[1,2,3,4,5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name="yld", description='yield of reaction', bounds=[0,100], is_objective=True) >>> strategy = SOBO(domain) >>> next_experiments = strategy.suggest_experiments(5)
Notes
- Gaussian Process (GP) model
GP: standard Gaussian Process
GP_MCMC: Gaussian Process with prior in hyperparameters
sparseGP: sparse Gaussian Process
warpedGP: warped Gaussian Process
InputWarpedGP: input warped Gaussian Process
RF: random forest (scikit-learn)
- Acquisition function type
EI: expected improvement
EI_MCMC: integrated expected improvement (requires GP_MCMC model) (https://dash.harvard.edu/bitstream/handle/1/11708816/snoek-bayesopt-nips-2012.pdf?sequence%3D1)
LCB: lower confidence bound
LCB_MCMC: integrated GP-Lower confidence bound (requires GP_MCMC model)
MPI: maximum probability of improvement
MPI_MCMC: maximum probability of improvement (requires GP_MCMC model)
LP: local penalization
ES: entropy search
This implementation uses the python package GPyOpt provided by the Machine Learning Group of the University of Sheffield.
Github repository: https://github.com/SheffieldML/GPyOpt
-
suggest_experiments(num_experiments=1, prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using GPyOpt single-objective Bayesian Optimization
- Parameters
num_experiments (int, optional) – The number of experiments (i.e., samples) to generate. Default is 1.
prev_res (
DataSet, optional) – Dataset with data from previous experiments of previous iteration. If no data is passed, then random sampling will be used to suggest an initial design.
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
MTBO¶
-
class
summit.strategies.MTBO(domain: summit.domain.Domain, pretraining_data: Optional[summit.utils.dataset.DataSet] = None, transform: Optional[summit.strategies.base.Transform] = None, task: int = 1, categorical_method: str = 'one-hot', **kwargs)[source]¶ Multitask Bayesian Optimisation
This strategy enables pre-training a model with past reaction data in order to enable faster optimisation.
- Parameters
domain (
Domain) – The domain of the optimizationpretraining_data (
DataSet, optional) – A DataSet with pretraining data. Must contain a metadata column named “task” that specfies the task for all data.transform (
Transform, optional) – A transform object. By default, no transformation will be done on the input variables or objectives.task (int, optional) – The index of the task being optimized. Defaults to 1.
categorical_method (str, optional) – The method for transforming categorical variables. Either “one-hot” or “descriptors”. Descriptors must be included in the categorical variables for the later.
Notes
This strategy is based on a paper from the NIPs2020 ML4Molecules workshop by Felton. See Swersky for more information on multitask Bayesian optimization.
References
- Felton
Felton, et al, in ML4 Molecules 2020 workshop.
- Swersky
Swersky et al., in NIPS Proceedings, 2013, pp. 2004–2012.
Examples
>>> from summit.benchmarks import MIT_case1, MIT_case2 >>> from summit.strategies import LHS, MTBO >>> from summit import Runner >>> # Get pretraining data >>> exp_pt = MIT_case1(noise_level=1) >>> lhs = LHS(exp_pt.domain) >>> conditions = lhs.suggest_experiments(10) >>> pt_data = exp_pt.run_experiments((conditions)) >>> pt_data[("task", "METADATA")] = 0 >>> # Use MTBO on a new mechanism >>> exp = MIT_case2(noise_level=1) >>> strategy = MTBO(exp.domain,pretraining_data=pt_data, categorical_method="one-hot",task=1) >>> r = Runner(strategy=strategy, experiment=exp, max_iterations=2) >>> r.run(progress_bar=False)
-
classmethod
load(filename)¶ Load a strategy from a JSON file
-
save(filename)¶ Save a strategy to a JSON file
-
suggest_experiments(num_experiments, prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using MTBO
- Parameters
num_experiments (int) – The number of experiments (i.e., samples) to generate
prev_res (
DataSet, optional) – Dataset with data from previous experiments. If no data is passed, then latin hypercube sampling will be used to suggest an initial design.
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
Examples
>>> from summit.benchmarks import MIT_case1, MIT_case2 >>> from summit.strategies import LHS, MTBO >>> from summit import Runner >>> # Get pretraining data >>> exp_pt = MIT_case1(noise_level=1) >>> lhs = LHS(exp_pt.domain) >>> conditions = lhs.suggest_experiments(10) >>> pt_data = exp_pt.run_experiments(conditions) >>> pt_data["task", "METADATA"] = 0 >>> # Use MTBO on a new mechanism >>> exp = MIT_case2(noise_level=1) >>> new_conditions = lhs.suggest_experiments(10) >>> data = exp.run_experiments(new_conditions) >>> data[("task", "METADATA")] = 1 >>> strategy = MTBO(exp.domain,pretraining_data=pt_data, categorical_method="one-hot",task=1) >>> res = strategy.suggest_experiments(1, prev_res=data)
ENTMOOT¶
-
class
summit.strategies.ENTMOOT(domain, transform=None, estimator_type=None, std_estimator_type=None, acquisition_type=None, optimizer_type=None, generator_type=None, initial_points=50, min_child_samples=5, **kwargs)[source]¶ Single-objective Bayesian optimization, using gradient-boosted trees instead of Gaussian processes, via ENTMOOT (ENsemble Tree MOdel Optimization Tool)
This is currently an experimental feature and requires Gurobipy to be installed.
- Parameters
domain (
Domain) – The Summit domain describing the optimization problem.transform (
Transform, optional) – A transform object. By default no transformation will be done on the input variables or objectives.estimator_type (string, optional) – The ENTMOOT base_estimator type. By default, Gradient-Boosted Regression
std_estimator_type (string, optional) – The ENTMOOT std_estimator By default, bounded data distance
acquisition_type (string, optional) – The acquisition function type from ENTMOOT. See notes for options. By default, Lower Confidence Bound.
optimizer_type (string, optional) – The optimizer used in ENTMOOT for maximization of the acquisition function. By default, sampling will be used.
generator_type (string, optional) – The method for generating initial points before a model can be trained. By default, uniform random points will be used.
initial_points (int, optional) – How many points to require before training models
min_child_samples (int, optional) – Minimum size of a leaf in tree models
Examples
>>> from summit.domain import * >>> from summit.strategies.entmoot import ENTMOOT >>> import numpy as np >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += CategoricalVariable(name='flowrate_a', description='flow of reactant a in mL/min', levels=[1,2,3,4,5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name="yld", description='yield of reaction', bounds=[0,100], is_objective=True) >>> # strategy = ENTMOOT(domain) >>> # next_experiments = strategy.suggest_experiments(5)
Notes
Estimator type can either by GBRT (Gradient-boosted regression trees) or RF (random forest from scikit-learn).
Acquisition function type can only be LCB (lower confidence bound).
Based on the paper from [Thebelt] et al.
-
gurobi_missing¶ Sets an initial points generator. Can be either - “random” for uniform random numbers, - “sobol” for a Sobol sequence, - “halton” for a Halton sequence, - “hammersly” for a Hammersly sequence, - “lhs” for a latin hypercube sequence, - “grid” for a uniform grid sequence
-
suggest_experiments(num_experiments=1, prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using ENTMOOT tree-based Bayesian Optimization
- Parameters
num_experiments (int, optional) – The number of experiments (i.e., samples) to generate. Default is 1.
prev_res (
DataSet, optional) – Dataset with data from previous experiments of previous iteration. If no data is passed, then random sampling will be used to suggest an initial design.
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
Reinforcement Learning¶
Reinforcement learning (RL) is distinct because it focuses on creating a custom policy for a particular problem instead of a model of the problem. In the case of reaction optimisation, the policy directly predicts the next experiment(s) should be given a history of past experiments. Policies are trained to maximise some sort of reward, such as achieving the maximum number of yield in as few experiments possible.
For more information about RL, see the book by Sutton and Barto or David Silver’s course.
-
class
summit.strategies.deep_reaction_optimizer.DRO(domain: summit.domain.Domain, transform: Optional[summit.strategies.base.Transform] = None, pretrained_model_config_path=None, model_size='standard', **kwargs)[source]¶ Deep Reaction Optimizer (DRO)
The DRO relies on a pretrained RL policy that can predict a next set of experiments given a set of past experiments. We suggest reading the notes below before using the DRO.
- Parameters
domain (
Domain) – A summit domain objecttransform (
Transform, optional) – A transform class (i.e, not the object itself). By default no transformation will be done the input variables or objectives.pretrained_model_config_path (string, optional) – Path to the config file of a pretrained DRO model (note that the number of inputs parameters should match the domain inputs) By default: a pretrained model will be used.
model_size (string, optional) – Whether the model (policy) has the same size as originally published by the developers of DRO (“standard”), or whether the model is bigger w.r.t. number of pretraining epochs, LSTM hidden size, unroll_length (“bigger”). Note that the pretraining can increase exponentially when changing these hyperparameters and the number of input variables, the number of epochs the each bigger model was trained can be found in the “checkpoint” file in the respective save directory. By default: “standard” (these models were all pretrained for 50 epochs)
-
xbest, internal state Best point from all iterations.
-
fbest, internal state Objective value at best point from all iterations.
-
param, internal state A dict containing: state of LSTM of DRO, last requested point, xbest, fbest, number of iterations (corresponding to the unroll length of the LSTM)
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import DRO >>> from summit.utils.dataset import DataSet >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> strategy = DRO(domain)
Notes
The DRO requires Tensorflow version 1, while all other parts of Summit use Tensorflow version 2. Therefore, we have created a Docker container for running DRO which has TFv1 installed. We also have an option in the pip package to install TFv1.
However, if you simply want to analyse results from a DRO run (i.e., use from_dict), then you will not get a tensorflow import error.
We have pretrained policies for domains with up to six continuous decision variables
For applying the DRO it is necessary to define reasonable bounds of the objective variable, e.g., yield in [0, 1], since the DRO normalizes the objective function values to be between 0 and 1.
The DRO is based on the paper in ACS Central Science by [Zhou].
References
- Zhou
Z. Zhou et al., ACS Cent. Sci., 2017, 3, 1337–1344. DOI: 10.1021/acscentsci.7b00492
-
suggest_experiments(prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using the Deep Reaction Optimizer
- Parameters
num_experiments (int, optional) – The number of experiments (i.e., samples) to generate. Default is 1.
prev_res (
DataSet, optional) – Dataset with data from previous experiments. If no data is passed, the DRO optimization algorithm will be initialized and suggest initial experiments.
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
Notes
Simplex¶
-
class
summit.strategies.neldermead.NelderMead(domain: summit.domain.Domain, transform: Optional[summit.strategies.base.Transform] = None, **kwargs)[source]¶ Nelder-Mead Simplex
A reimplementation of the Nelder-Mead Simplex method adapted for sequential calls. This includes adaptions in terms of reflecting points, dimension reduction and dimension recovery proposed by Cortes-Borda et al.
- Parameters
domain (
Domain) – The domain of the optimizationtransform (
Transform, optional) – A transform object. By default no transformation will be done on the input variables or objectives.random_start (bool, optional) – Whether to start at a random point or the value specified by x_start
adaptive (bool, optional) – Adapt algorithm parameters to dimensionality of problem. Useful for high-dimensional minimization. Default is False.
x_start (array_like of shape (1, N), optional) – Initial center point of simplex Default: empty list that will initialize generation of x_start as geoemetrical center point of bounds Note that x_start is ignored when initial call of suggest_exp contains prev_res and/or prev_param
dx (float, optional) – Parameter for stopping criterion: two points are considered to be different if they differ by at least dx(i) in at least one coordinate i. Default is 1E-5.
df (float, optional) – Parameter for stopping criterion: two function values are considered to be different if they differ by at least df. Default is 1E-5.
Notes
This is inspired by the work by [Cortés-Borda]. Implementation partly follows the Nelder-Mead Simplex implementation in scipy-optimize
After the initialisation, the number of suggested experiments depends on the internal state of Nelder Mead. Usually the algorithm requests 1 point per iteration, e.g., a reflection. In some cases it requests more than 1 point, e.g., for shrinking the simplex.
References
- Cortés-Borda
Cortés-Borda, D.; Kutonova, K. V.; Jamet, C.; Trusova, M. E.; Zammattio, F.; Truchet, C.; Rodriguez-Zubiri, M.; Felpin, F.-X. Optimizing the Heck–Matsuda Reaction in Flow with a Constraint-Adapted Direct Search Algorithm. Organic ProcessResearch & Development 2016,20, 1979–1987
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import NelderMead >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[0, 1]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0, 1]) >>> domain += ContinuousVariable(name="yld", description='relative conversion to xyz', bounds=[0,100], is_objective=True, maximize=True) >>> strategy = NelderMead(domain) >>> next_experiments = strategy.suggest_experiments() >>> print(next_experiments) NAME temperature flowrate_a strategy TYPE DATA DATA METADATA 0 0.500 0.500 Nelder-Mead Simplex 1 0.625 0.500 Nelder-Mead Simplex 2 0.500 0.625 Nelder-Mead Simplex
-
round(x, bounds, dx)[source]¶ function x = round(x, bounds, dx)
A point x is projected into the interior of [u, v] and x[i] is rounded to the nearest integer multiple of dx[i].
Input: x vector of length n bounds matrix of length nx2 such that bounds[:,0] < bounds[:,1] dx float
Output: x projected and rounded version of x
-
suggest_experiments(prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using Nelder-Mead Simplex method
- Parameters
prev_res (summit.utils.data.DataSet, optional) – Dataset with data from previous experiments. If no data is passed, the Nelder-Mead optimization algorithm will be initialized and suggest initial experiments.
- Returns
next_experiments – A Dataset object with the suggested experiments by Nelder-Mead Simplex algorithm
- Return type
Notes
After the initialisation, the number of suggested experiments depends on the internal state of Nelder Mead. Usually the algorithm requests 1 point per iteration, e.g., a reflection. In some cases it requests more than 1 point, e.g., for shrinking the simplex. Thus, there is no num_experiments keyword argument.
Random¶
Random¶
-
class
summit.strategies.random.Random(domain: summit.domain.Domain, transform: Optional[summit.strategies.base.Transform] = None, random_state: Optional[numpy.random.mtrand.RandomState] = None, **kwargs)[source]¶ Random strategy for experiment suggestion
- Parameters
domain (summit.domain.Domain) – A summit domain object
random_state (np.random.RandomState`) – A random state object to seed the random generator
-
domain¶
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import Random >>> import numpy as np >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> strategy = Random(domain, random_state=np.random.RandomState(3)) >>> strategy.suggest_experiments(5) NAME temperature flowrate_a flowrate_b strategy TYPE DATA DATA DATA METADATA 0 77.539895 0.458517 0.111950 Random 1 85.407391 0.150234 0.282733 Random 2 64.545237 0.182897 0.359658 Random 3 75.541380 0.120587 0.211395 Random 4 94.647348 0.276324 0.370502 Random
Notes
Descriptors variables are selected randomly as if they were discrete variables instead of sampling evenly in the continuous space.
-
suggest_experiments(num_experiments: int, **kwargs) → summit.utils.dataset.DataSet[source]¶ Suggest experiments for a random experimental design
- Parameters
num_experiments (int) – The number of experiments (i.e., samples) to generate
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
Latin Hypercube Sampling¶
-
class
summit.strategies.random.LHS(domain: summit.domain.Domain, transform: Optional[summit.strategies.base.Transform] = None, random_state: Optional[numpy.random.mtrand.RandomState] = None, categorical_method: Optional[str] = None)[source]¶ Latin hypercube sampling (LHS) strategy for experiment suggestion
LHS samples evenly throughout the continuous part of the domain, which can result in better data for model training.
- Parameters
domain (summit.domain.Domain) – A summit domain object
random_state (np.random.RandomState`) – A random state object to seed the random generator
categorical_method (str, optional) – The method for transforming categorical variables. Either “one-hot” or “descriptors”. Descriptors must be included in the categorical variables for the later.
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import LHS >>> import numpy as np >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> strategy = LHS(domain, random_state=np.random.RandomState(3)) >>> strategy.suggest_experiments(5) NAME temperature flowrate_a flowrate_b strategy TYPE DATA DATA DATA METADATA 0 95.0 0.46 0.38 LHS 1 65.0 0.14 0.14 LHS 2 55.0 0.22 0.30 LHS 3 85.0 0.30 0.46 LHS 4 75.0 0.38 0.22 LHS
Notes
LHS was first introduced by [McKay] and coworkers in 1979. We rely on the implementation from pyDoE2.
Our version randomly selects a categorical variable if no descriptors are available. If descriptors are available it samples in the continuous space and then chooses the closest point by Euclidean distance.
References
- McKay
R.J. Beckman et al., Technometrics, 1979, 21, 239–245.
-
suggest_experiments(num_experiments, criterion='center', exclude=[], **kwargs) → summit.utils.dataset.DataSet[source]¶ Generate latin hypercube intial design
- Parameters
num_experiments (int) – The number of experiments (i.e., samples) to generate
criterion (str, optional) – The criterion used for the LHS. Allowable values are “center” or “c”, “maximin” or “m”, “centermaximin” or “cm”, and “correlation” or “corr”. Default is center.
exclude (array like, optional) – List of variable names that should be excluded from the design. Default is None.
- Returns
next_experiments – A Dataset object with the suggested experiments
- Return type
DataSet
Other¶
SNOBFIT¶
-
class
summit.strategies.snobfit.SNOBFIT(domain: summit.domain.Domain, **kwargs)[source]¶ Stable Noisy Optimization by Branch and Fit (SNOBFIT)
SNOBFIT is designed to quickly optimise noisy functions.
- Parameters
domain (
Domain) – The domain of the optimizationtransform (
Transform, optional) – A transform object. By default no transformation will be done on the input variables or objectives.probability_p (float, optional) – The probability p that a point of class 4 is generated, i.e., higher p leads to more exploration. Default is 0.5.
dx_dim (float, optional) – only used for the definition of a new problem: two points are considered to be different if they differ by at least dx(i) in at least one coordinate i. Default is 1E-5.
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import SNOBFIT >>> from summit.utils.dataset import DataSet >>> import pandas as pd >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[0, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0, 1]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.9]) >>> domain += ContinuousVariable(name="yld", description='relative conversion to xyz', bounds=[0,100], is_objective=True, maximize=True) >>> d = {'temperature': [50,40,70,30], 'flowrate_a': [0.6,0.3,0.2,0.1], 'flowrate_b': [0.1,0.3,0.2,0.1], 'yld': [0.7,0.6,0.3,0.1]} >>> df = pd.DataFrame(data=d) >>> initial = DataSet.from_df(df) >>> strategy = SNOBFIT(domain) >>> next_experiments = strategy.suggest_experiments(5, initial)
Notes
SNOBFIT was created by [Huyer] et al. This implementation is based on the python reimplementation [SQSnobFit] of the original MATLAB code by [Neumaier].
Note that SNOBFIT sometimes returns more experiments than requested when the number of experiments request is small (i.e., 1 or 2). This seems to be a general issue with the algorithm instead of the specific implementation used here.
References
- Huyer
W. Huyer et al., ACM Trans. Math. Softw., 2008, 35, 1–25. DOI: 10.1145/1377612.1377613.
- SQSnobFit
Lavrijsen, W. SQSnobFit https://pypi.org/project/SQSnobFit/
- Neumaier
-
snobfit(x, f, config, dx=None, prev_param=None)[source]¶ The following snobfit code was copied and modified from the SQSnobFit package and was originally published by Wim Lavrijsen. The SQSnobFit package includes a python version of SNOBFIT which was originally published by A. Neumaier.
- Copyright of SNOBFIT (v2.1):
Neumaier, University of Vienna
Website: https://www.mat.univie.ac.at/~neum/software/snobfit/
- Copyright of SQSnobfit (v0.4.2)
UC Regents, Berkeley
Website: https://pypi.org/project/SQSnobFit/
-
suggest_experiments(num_experiments=1, prev_res: Optional[summit.utils.dataset.DataSet] = None, **kwargs)[source]¶ Suggest experiments using the SNOBFIT method
- Parameters
num_experiments (int, optional) – The number of experiments (i.e., samples) to generate. Default is 1.
prev_res (summit.utils.data.DataSet, optional) – Dataset with data from previous experiments. If no data is passed, the SNOBFIT optimization algorithm will be initialized and suggest initial experiments.
- Returns
next_experiments – A Dataset object with the suggested experiments by SNOBFIT algorithm
- Return type
Full Factorial¶
-
class
summit.strategies.factorial_doe.FullFactorial(domain: summit.domain.Domain, transform: Optional[summit.strategies.base.Transform] = None, **kwargs)[source]¶ Full factorial DoE Strategy for full factorial design of experiments in all decision variables.
- Parameters
domain (
Domain) – The Summit domain describing the optimization problem.
Examples
>>> from summit.domain import Domain, ContinuousVariable >>> from summit.strategies import FullFactorial >>> import numpy as np >>> domain = Domain() >>> domain += ContinuousVariable(name='temperature', description='reaction temperature in celsius', bounds=[50, 100]) >>> domain += ContinuousVariable(name='flowrate_a', description='flow of reactant a in mL/min', bounds=[0.1, 0.5]) >>> domain += ContinuousVariable(name='flowrate_b', description='flow of reactant b in mL/min', bounds=[0.1, 0.5]) >>> levels = dict(temperature=[50,100], flowrate_a=[0.1,0.5], flowrate_b=[0.1,0.5]) >>> strategy = FullFactorial(domain) >>> strategy.suggest_experiments(levels) NAME temperature flowrate_a flowrate_b strategy TYPE DATA DATA DATA METADATA 0 50.0 0.1 0.1 FullFactorial 1 100.0 0.1 0.1 FullFactorial 2 50.0 0.5 0.1 FullFactorial 3 100.0 0.5 0.1 FullFactorial 4 50.0 0.1 0.5 FullFactorial 5 100.0 0.1 0.5 FullFactorial 6 50.0 0.5 0.5 FullFactorial 7 100.0 0.5 0.5 FullFactorial
Notes
We rely on the implementation from pyDoE2.
-
suggest_experiments(levels_dict, **kwargs) → summit.utils.dataset.DataSet[source]¶ Suggest experiments for a full factorial experimental design
- Parameters
levels_dict (dict) – A dictionary with the number of levels for each variable. Keys are the variable names and values are arrays with the values of each level.
- Returns
A Dataset object with the random design
- Return type
ds